Unlocking Advanced Metadata Extraction with the New DBT API in Dagster
Applying the recent DBT API improvements of Dagster in action

overview
As data engineering professionals, staying at the forefront of innovative tools and techniques is crucial for optimizing our workflows. Recently, Dagster, the powerful data orchestration framework, introduced an enhanced DBT integration, bringing a wealth of opportunities for seamless collaboration and advanced metadata extraction. In this blog post, we will explore the capabilities of the new DBT API in Dagster and demonstrate its usage with a concrete example of metadata extraction and integration into the data catalog Open Metadata.
the new API
With the recent upgrades dagster makes its DBT API more composable for the end user. However, a couple of changes are relevant to note
DBT target
Dagster is unifying the resource handlign of DBT + dagster resources (removing the old dagster resource for DBT here) to configure all DBT related parts with the DBT native means.
For example you could retrieve the desired target from an ENV variable and then feed it to the DBT CLI to support i.e. branch deployments in dagster.
target = os.environ.get("DBT_TARGET", "dev")
context.log.debug(f"Running dbt with target {target}")
# in the definitions
from dagster_dbt import DbtCliResource
dbt = DbtCliResource(
project_dir="/path/to/dbt/project",
global_config_flags=["--no-use-colors"],
profile="jaffle_shop",
target="dev",
)
dbt_cli_task = dbt.cli(
["run", "--profiles-dir", DBT_PROFILES_DIR],
manifest=manifest,
context=context,
)
retrieving metadata from DBT
Since DBT 1.4 dagsters DBT integration is no longer blocking. However, if you need to retrieve advanced metadata from DBTs output files i.e. record count of the affected rows then a blocking execution of DBT is required:
dbt_cli_task.wait()
Then the assets can be retrieved:
run_results = dbt_cli_task.get_artifact("run_results.json")
executed_manifest = dbt_cli_task.get_artifact("manifest.json")
Finally, the relevant meta information can be extracted from them:
# Then, we can use the run results to add metadata to the outputs.
for event in dbt_cli_task.stream_raw_events():
for dagster_event in event.to_default_asset_events(manifest=manifest):
if isinstance(dagster_event, Output):
event_node_info = event.event["data"]["node_info"]
output_name = dagster_event.output_name
result = results_by_output_name[output_name]
rows_affected: Optional[int] = result["adapter_response"].get(
"rows_affected"
)
rows_affected_metadata = (
{"rows_affected": rows_affected} if rows_affected else {}
)
node = manifest_by_output_name[output_name]
compiled_sql: Optional[str] = node.get("compiled_code")
compiled_sql_metadata = (
{"compiled_sql": MetadataValue.md(compiled_sql)}
if compiled_sql
else {}
)
context.add_output_metadata(
metadata={
**rows_affected_metadata,
**compiled_sql_metadata,
},
output_name=output_name,
)
yield dagster_event
Notice, Dagster and DBT use different primary keys (asset key vs. unique_id), a lookup table must be created before you can access the metadata.
results_by_output_name = {
output_name_fn({"unique_id": result["unique_id"]}): result
for result in run_results["results"]
}
manifest_by_output_name = {
output_name_fn({"unique_id": unique_id}): node
for unique_id, node in executed_manifest["nodes"].items()
}
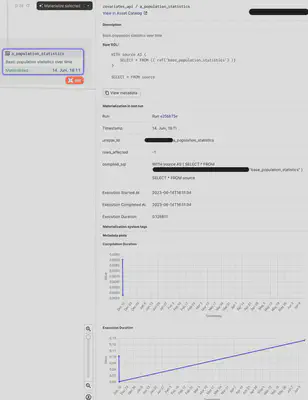
The extracted metrics can then directly be visualized in Dagster:

When the asset is selected in dagit, the UI is directly showing all the metadata and plots at a glance:
openmetadata DBT integration with dagster
For more advanced data governance/data management usecases a dedicated data catalog is required. One such tool is Open Metadata. Open Metadata is a data catalog. It has a great DBT integration. There is no first class support for a dagster ingestion process yet: https://github.com/open-metadata/OpenMetadata/issues/7762 However, the DBT CLI of Open Metadata can easily be accessed from dagster.
You need to configure a file similar to:
source:
type: dbt
serviceName: "my_db${DEPLOYMENT_NAME}"
sourceConfig:
config:
type: DBT
dbtConfigSource:
dbtCatalogFilePath: /path/to/dbt/target/catalog.json
dbtManifestFilePath: /path/to/dbt/target/manifest.json
dbtRunResultsFilePath: /path/to/dbt/target/run_results.json
sink:
type: metadata-rest
config: {}
workflowConfig:
openMetadataServerConfig:
hostPort: $OPEN_METADATA_URL
authProvider: openmetadata
securityConfig:
jwtToken: $OPEN_METADATA_INGESTION_TOKEN
It can then be ingested using either the bare CLI like:
metadata ingest -c /path/to/recipe_dbt.yml
or most likely better using the native python workflow:
# NOTICE: partial pseudocode only
from metadata.ingestion.api.workflow import Workflow
workflow = Workflow.create(workflow_config)
workflow.execute()
workflow.raise_from_status()
workflow.print_status()
workflow.stop()
Before you can shedule the DBT ingest to Open Metadata you have to retrieve the DBT artifacts. As discussed before, the artifacts i.e. catalog, tests, … have to be created. Then they can be passed along.
However, dagster will create a dynamic path to support running multiple instances of the pipeline in parallel. The OM process needs to be fed with a suitable path to a JSON file in order to execute successfully:
# NOTICE: partial pseudocode only
dbt_cli_task.wait()
executed_manifest = dbt_cli_task.get_artifact("manifest.json")
executed_catalog = dbt_cli_task.get_artifact("catalog.json")
write_json_to_file(executed_manifest, "om_dbt_this_dynamic_path/target/manifest.json")
write_json_to_file(executed_catalog, "om_dbt_this_dynamic_path/target/catalog.json")
dbt_cli_task = dbt.cli(
["test", "--profiles-dir", DBT_PROFILES_DIR],
manifest=manifest,
)
# Run the task, but don't yield events (and block execution).
_ = list(dbt_cli_task.stream_raw_events())
run_results = dbt_cli_task.get_artifact("run_results.json")
write_json_to_file(run_results, "MWF_dbt/target/run_results.json")
summary
The new DBT API integration in Dagster offers data engineering professionals an exciting opportunity to unlock advanced metadata extraction and integration capabilities. By harnessing the power of the DBT API, we can enhance data governance, improve collaboration, and streamline data cataloging processes. Start exploring the new DBT API in Dagster today and elevate your data engineering workflows to new heights.
The full E2E code example for the new API of DBT can be found on github: https://gist.github.com/geoHeil/a7ee774e65b597e9ac534e678cf5ecfc.
The image was generated using firefly by adobe. Parts of the text were neatly generated by chatgpt.
See also https://github.com/dagster-io/dagster/discussions/14477 for more details